MRI Preprocessing Resources
- Emma Garrison
- Dec 26, 2025
- 7 min read
For my Master's project, I was required to preprocess Magnetic Resonance Imaging (MRI) data. Now, this is actually much trickier than I originally expected. So, I thought I would share some resources and tips that helped me!
I preprocessed both functional Magnetic Resonance Imaging (fMRI) and Diffusion Tensor Imaging (DTI) data for the formation of networks. This is not a fully comprehensive guide, but hopefully it could give you a head start when beginning your journey.
Background
For me personally, MRI machines are not particularly intuitive. This subject was difficult to learn even with the leg up I received from knowing physics. While I don't think it is necessary for everyone that is preprocessing MRI data to understand all the ins and outs of the MRI machine, I found as I was working that I could not understand some of the steps without understanding the base concepts. I am very pleased that I learned this information though I have been told that is was "a bit much" for a master's thesis that was not primarily about preprocessing. However, I do find that I am able to speak more plainly about the assumptions and measurement details in these kind of data because I have taken the time to understand the background. So, if you are as curious (and as crazy) as I am, I think you will find these resources to be incredibly helpful to understand the background of MRI machines!
Radiology tutorials
First, and probably to most useful, are the Radiology Tutorials from Dr. Michael Nel. He does a great job breaking down the concepts in his MRI Physics playlist.
I particularly liked the consistency of his timing diagrams, the clarity of his animations, and the detail in his explanations. I, unfortunately, must recommend that you watch the series from the beginning because skipping around can be somewhat confusing (at least to me). However, once you have the basics you can start applying your understanding to your work immediately.
A couple things that might help those that are not Radiologist. Different kinds of MRI images are created from different MRI sequences. These are the patterns of radiofrequency pulses and readouts that occur during imaging. These sequences are discussed in Dr. Nel's videos. However, I did not know which sequences were associated with fMRI and DTI before I started. Once you have a base understanding of the baseline MRI sequences, you can watch the video on Fast Pulse Sequences where you were learn about Echo Planar Imaging which are used in fMRI and DTI. He also has a video specifically on DTI called Diffusion Weighted Imaging. These videos will help you understand the sequences that will be used in your dataset.
vocabulary and basics
The key to preprocessing understanding your MRI Protocols. These should be readily available in your dataset; however sometimes you have to go looking to find them!
Here are a couple of terms that you might need to understand.
Volume
This is just one full scan of the brain. The brain is scanned in slices and by deciphering spatial encoding information. Volume just refers to the whole brain scan! So in fMRI and DTI, the time series is made up of a bunch of volumes (sometimes you will see this language in graphs).
TR - Time to Repetition
This is the time between repeated radiofrequency pulses and the time is takes to scan 1 volume.
TE - Time to Echo
This is the time between the radiofrequency pulse and the readout of the signal. This is when the machine measures the signal.
Multiband - Simultaneous Multi-Slice (SMS)
This refers to scans where multiple slices of the brain are scanned at once. This reduced the TR value and usually improved both the temporal and spatial resolution of the scan.
Here are timing diagrams I created showing the difference between a Basic Scan (red) and a Multiband Scan (blue).
Basic Scan

Multiband Scan

Remember that the whole brain (or Volume) is scanned in one TR.
Multiband Factor
This is the number that refers to the number of volumes that are scanned simultaneously in a SMS scan. In the diagram above the multiband factor is 8.
Interleaved Acquisition
This refers to the most common acquisition protocol in which slices are scanned in an interwoven pattern to reduce bias across the time of TR.
For example, instead of scanning slices A,B,C,D,E,F the interleaved acquisition would scan slices A,D,B,E,C,F.
Other Helpful Videos
Here are a few more YouTube videos that might help you understand the science behind MRIs as well as the underlying physiology of the brain! I may add to this list if I come across any particularly good videos moving forward.
MRI
https://www.youtube.com/watch?v=jLnuPKhKXVM - In this video from Johns Hopkins Medicine, Dr. Gomez gives a great explanation of the general Spin Echo Sequence of MRI. I wished there were a whole series for the other sequences!
fMRI
https://www.youtube.com/watch?v=4UOeBM5BwdY - Here is a very quick and visual explanation of the physiology of the BOLD signal measured in fMRI from the Wellcome Center for Human Neuroimaging
https://www.youtube.com/watch?v=GDx40b-FZiQ&t=563s - In this video, Doctor Klioze goes into greater detail about hemoglobin and uses the language that you will encounter throughout preprocessing.
DTI
https://www.youtube.com/watch?v=twsV81UFFcE - Here Doctor Klioze goes into greater detail about the mathematics of DTI. This is helpful for understanding the tractography algorithm during preprocessing.
https://www.youtube.com/watch?v=nZU_5TH1CWs - In this video from the Cognitive Neuroscience Compendium, Dr. Alexander uses real MRI images to demonstrate the concepts.
Preprocessing Steps
Andy's Brain book
The resources that is the most useful for laying out the preprocessing steps is Andy's Brain Book where Dr. Andy Jahn at the University of Michigan puts together step by step guides for preprocessing. This is a great reference that explains how and why steps are taken.

There is a fMRI Preprocessing Guide and a DTI Preprocessing Guide. These provide detailed instructions for preprocessing. I will say that if you are looking to write a preprocessing script for a large dataset there are a few more steps of work that you need to do. The fMRI Guide in particular relies on the FSL GUI rather than a script. However, this is a great way to start to understand the steps that are taken during preprocessing and give you the language to read research papers regarding the merits and parameters of these steps.
Tools and Documentation
There are several tools that one can use for preprocessing each with their own quirks and challenges. I have included here the tools that I used; however, this is not a comprehensive list.
SPM - Statistical Parametric Mapping
I used this software package for fMRI preprocessing. This can be scripted in MATLAB so that you can preprocess all your data. Here is there preprocessing documentation.
FSL - FMRIB Software Library
I used this for steps in both the fMRI and DTI preprocessing. It is very important to note that this program will not work on a Windows machine so you will need access to a Linux terminal if you wish to run this on Windows. Here is FSL's documentation.
MRtrix3
I used this tool for DTI preprocessing as described in Andy's Brain Book. I simply wrote a MATLAB script as a series of command line prompts in order to systematize this. The documentation is very helpful for this one because there are so many parameters.
In general, I had to learn or review many things before I had scripts that were anywhere near operational. Getting comfortable with writing scripts, managing files from scripts, working in the command line, accessing a Linux terminal (I was working on windows), and effectively using documentation will be essential for executing the preprocessing steps.
Visualizations and checks
I think it is important throughout preprocessing to sanity check what you are doing. Essentially, making sure the images and results look how you (or a trusted advisor) would expect. This has the added benefit of giving you visualizations to use to report on your preprocessing. My recommendation is to take every opportunity to visualize after each step.
I have a few tools that are helpful for visualizations; however, many steps in this process require different kinds of visualizations many of which I did in MATLAB. Unfortunately, this process is quite bespoke and visualization is one of the most difficult steps.
Tools and Documentation
mrview
This is a command in MRtrix3 which can help you visualize the data as you go and it will have the same documentation as MRtrix3.
SPM functions
Check your SPM functions because a few have built-in visualizations that you can turn on. Check out the SPM documentation for more details.
imshow
This, and related fuctions, can create brain images in MATLAB directly and has documentation here. I would describe this as a pain, but it is great is you want to put visualizations into your script.
FSL
I know that FSL has visualization capabilities though I didn't use them personally mostly because I was on a Windows machine.
MRIcron
This is tool that has a useful GUI and you can create various images of brain data. I find the documentation for this a little confusing, but the community questions in the forum are very useful!
3D Slicer
This tool similarly has a useful GUI for brain images. There are a lot of options for colors, layers, etc. This tool does a lot so it can be challenging to find what you are looking for in the documentation, but I was always able to find it.
BrainNet Viewer
This tool is for visualizing the resulting brain networks. I was creating networks in my project so I found this tool very useful. Similar to MRIcon, the forum is perhaps more helpful than the documentation.
I hope that these are useful for visualizations. I do think that creating visualizations can feel like a very straightforward step, but I found that is was not, especially because certain steps require different kinds of graphs. Below are the fMRI preprocessing graphics that I created for my master's thesis!
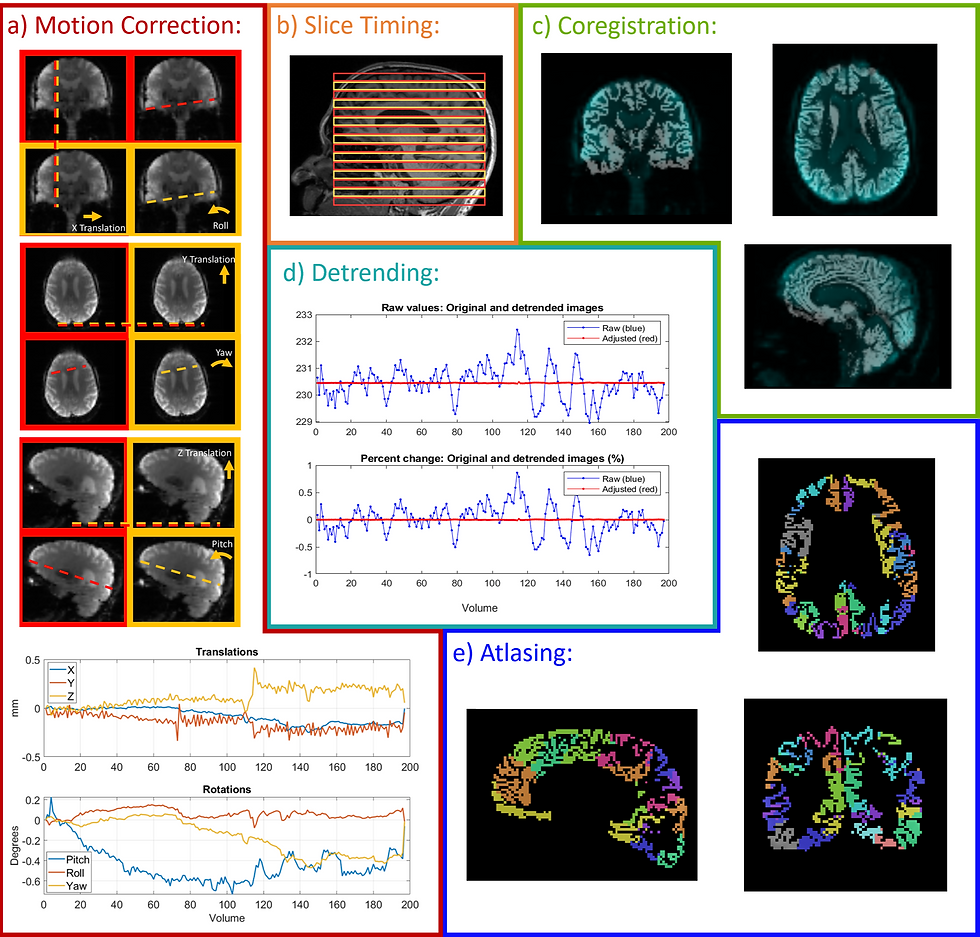

Conclusions
I am currently working on including some of my work in this area into my github, especially regarding the creation of visualizations inside the script itself. Additionally, I've been creating a resource for other students trying to understand MRI machines and preprocessing so that others do not have to search as much as I did. This resource list is just a small portion of that resource development; hopefully, it will be of some use to those working to preprocess MRI data!



Comments